What is Scientific Programming?
Scientific programming deals with solving scientific problems with the help of parallel/ cluster computers, so as to obtain results more quickly and accurately. Computers have long been used for solving complex scientific problems — however, advancements in computer science and hardware technologies over the years have also allowed students and academicians to play around with robust scientific computation tools. Scientific programming using a cluster of computers joins computational powers of the compute nodes to provide a more combined computational power.
Hands-on Scientific Computing Essentials
This is the first ever hands-on scientific programming course that uses the High Performance Computing (HPC) systems software stack: Slurm, PBS Pro, OpenMP, MPI and CUDA. The goal main of this course is to introduce you with the HPC systems and its software stack. This course has been specially designed to enable you to utilize parallel & distributed programming and computing resources to accelerate the solution of a complex problem with the help of HPC systems and Supercomputers. You can then use your knowledge in Machine learning, Deep learning, Data Sciences, Big data and so on.
You will learn
- Scientific programming in HPC clusters computers and is benefits, Supercomputing history and examples.
- Components of a High Performance Systems (HPC) cluster, Properties of Login node(s), Compute node(s), Master node(s), Storage node(s), HPC networks and so on.
- Introduction to PBS, PBS basic commands, PBS
qsub, PBSqstat, PBSqdelcommand, PBSqalter, PBS job states, PBS variables, PBS interactive jobs, PBS arrays, PBS Matlatb example - Introduction to Slurm, Slurm commands, A simple Slurm job, Slurm distrbuted MPI and GPU jobs, Slurm multi-threaded OpenMP jobs, Slurm interactive jobs, Slurm array jobs, Slurm job dependencies
- OpenMP basics, Open MP - clauses, worksharing constructs, OpenMP- Hello world!, reduction and parallel
for-loop, section parallelization, vector addition, MPI - hello world! send/ receive andping-pong - Friendly guide to the GPUs - graphics processing units, GPU Programming - CUDA, CUDA - hello world and so on!
Requirements
It is a self-learning course with all Linux environtments provided.
Foundations of C/C++
Virtual cluster
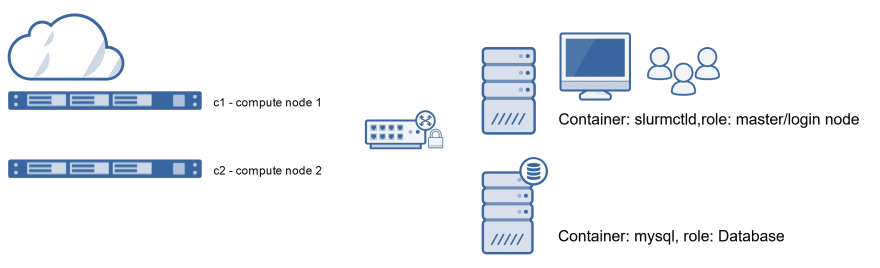
For this course, we created a small private virtual cluster with Docker technology. It has the following components:
- slurmctld: master node/ login node
- mysql and slurmdbd: database nodes
- c1: compute node 1
- c2: compute node 2
See further details: How do we deploy a virtual cluster for the course "Scientific Computing Essentials"?
Outcome
- Hands-on learning capability of Scientific computing
- HPC system's basic components
- HPC software stack
- HPC job schedulers and batch systems (Slurm and PBS Pro)
- Introduction to parallel programming concepts: Open MP and MPI
- GPU programming: CUDA
Points to note
We initially released this course on the Educative, but the platform do not yet support the relevant playgrounds (e.g., CUDA, virutal HPC, MPI and so on). Therefore re-published at the Scientific Programming School, which is a specialized platform for scientific coding. It's much better and interactive! Also forget to mention that the course is now FREE!
Why wait?
Scientific Computing Essentials @ Scientific Programming School



Top comments (0)