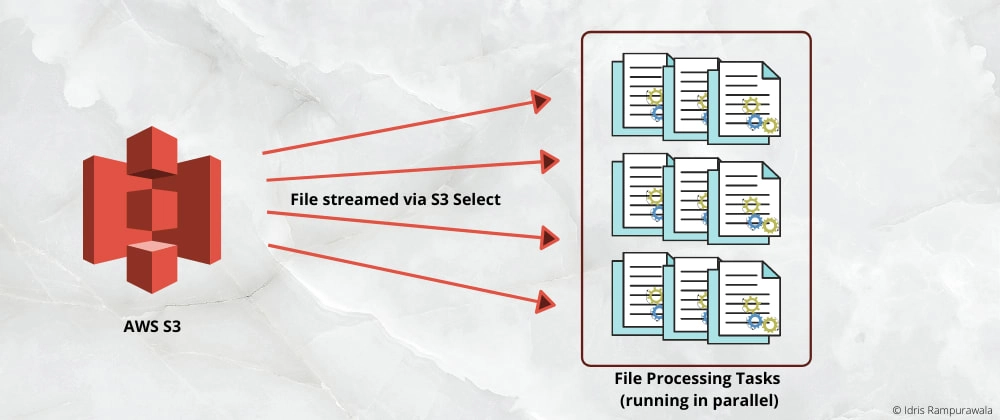
In my last post, we discussed achieving the efficiency in processing a large AWS S3 file via S3 select. The processing was kind of sequential and it might take ages for a large file. So how do we parallelize the processing across multiple units? 🤔 Well, in this post we gonna implement it and see it working!
📝 I highly recommend checking out my last post on streaming S3 file via S3-Select to set the context for this post.
I always like to break down a problem into the smaller pieces necessary to solve it (analytical approach). Let's try to solve this in 3 simple steps:
1. Find the total bytes of the S3 file
Very similar to the 1st step of our last post, here as well we try to find file size first.
The following code snippet showcases the function that will perform a HEAD request on our S3 file and determines the file size in bytes.
# core/utils.py
def get_s3_file_size(bucket: str, key: str) -> int:
"""Gets the file size of S3 object by a HEAD request
Args:
bucket (str): S3 bucket
key (str): S3 object path
Returns:
int: File size in bytes. Defaults to 0 if any error.
"""
aws_profile = current_app.config.get('AWS_PROFILE_NAME')
s3_client = boto3.session.Session(profile_name=aws_profile).client('s3')
file_size = 0
try:
response = s3_client.head_object(Bucket=bucket, Key=key)
if response:
file_size = int(response.get('ResponseMetadata').get('HTTPHeaders').get('content-length'))
except ClientError:
logger.exception(f'Client error reading S3 file {bucket} : {key}')
return file_size
2. Create a celery task to process a chunk
Here, we would define a celery task to process a file chunk (which will be executed in parallel later). The overall processing here will look like this:
- Receive the
startandend bytesof this chunk as an argument - Fetch this part of the S3 file via S3-Select and store it locally in a temporary file (as CSV in this example)
- Read this temporary file and perform any processing required
- Delete this temporary file
📝 I term this task as a file chunk processor. It processes a chunk from a file. Running multiple of these tasks completes the processing of the whole file.
# core/tasks.py
@celery.task(name='core.tasks.chunk_file_processor', bind=True)
def chunk_file_processor(self, **kwargs):
""" Creates and process a single file chunk based on S3 Select ScanRange start and end bytes
"""
bucket = kwargs.get('bucket')
key = kwargs.get('key')
filename = kwargs.get('filename')
start_byte_range = kwargs.get('start_byte_range')
end_byte_range = kwargs.get('end_byte_range')
header_row_str = kwargs.get('header_row_str')
local_file = filename.replace('.csv', f'.{start_byte_range}.csv')
file_path = path.join(current_app.config.get('BASE_DIR'), 'temp', local_file)
logger.info(f'Processing {filename} chunk range {start_byte_range} -> {end_byte_range}')
try:
# 1. fetch data from S3 and store it in a file
store_scrm_file_s3_content_in_local_file(
bucket=bucket, key=key, file_path=file_path, start_range=start_byte_range,
end_range=end_byte_range, delimiter=S3_FILE_DELIMITER, header_row=header_row_str)
# 2. Process the chunk file in temp folder
id_set = set()
with open(file_path) as csv_file:
csv_reader = csv.DictReader(csv_file, delimiter=S3_FILE_DELIMITER)
for row in csv_reader:
# perform any other processing here
id_set.add(int(row.get('id')))
logger.info(f'{min(id_set)} --> {max(id_set)}')
# 3. delete local file
if path.exists(file_path):
unlink(file_path)
except Exception:
logger.exception(f'Error in file processor: {filename}')
3. Execute multiple celery tasks in parallel
This is the most interesting step in this flow. We will create multiple celery tasks to run in parallel via Celery Group.
Once we know the total bytes of a file in S3 (from step 1), we calculate start and end bytes for the chunk and call the task we created in step 2 via the celery group. The start and end bytes range is a continuous range of file size. Optionally, we can also call a callback (result) task once all our processing tasks get completed.
# core/tasks.py
@celery.task(name='core.tasks.s3_parallel_file_processing', bind=True)
def s3_parallel_file_processing_task(self, **kwargs):
""" Creates celery tasks to process chunks of file in parallel
"""
bucket = kwargs.get('bucket')
key = kwargs.get('key')
try:
filename = key
# 1. Check file headers for validity -> if failed, stop processing
desired_row_headers = (
'id',
'name',
'age',
'latitude',
'longitude',
'monthly_income',
'experienced'
)
is_headers_valid, header_row_str = validate_scrm_file_headers_via_s3_select(
bucket=bucket,
key=key,
delimiter=S3_FILE_DELIMITER,
desired_headers=desired_row_headers)
if not is_headers_valid:
logger.error(f'{filename} file headers validation failed')
return False
logger.info(f'{filename} file headers validation successful')
# 2. fetch file size via S3 HEAD
file_size = get_s3_file_size(bucket=bucket, key=key)
if not file_size:
logger.error(f'{filename} file size invalid {file_size}')
return False
logger.info(f'We are processing {filename} file about {file_size} bytes :-o')
# 2. Create celery group tasks for chunk of this file size for parallel processing
start_range = 0
end_range = min(S3_FILE_PROCESSING_CHUNK_SIZE, file_size)
tasks = []
while start_range < file_size:
tasks.append(
chunk_file_processor.signature(
kwargs={
'bucket': bucket,
'key': key,
'filename': filename,
'start_byte_range': start_range,
'end_byte_range': end_range,
'header_row_str': header_row_str
}
)
)
start_range = end_range
end_range = end_range + min(S3_FILE_PROCESSING_CHUNK_SIZE, file_size - end_range)
job = (group(tasks) | chunk_file_processor_callback.s(data={'filename': filename}))
_ = job.apply_async()
except Exception:
logger.exception(f'Error processing file: {filename}')
@celery.task(name='core.tasks.chunk_file_processor_callback', bind=True, ignore_result=False)
def chunk_file_processor_callback(self, *args, **kwargs):
""" Callback task called post chunk_file_processor()
"""
logger.info('Callback called')
# core/utils.py
def store_scrm_file_s3_content_in_local_file(bucket: str, key: str, file_path: str, start_range: int, end_range: int,
delimiter: str, header_row: str):
"""Retrieves S3 file content via S3 Select ScanRange and store it in a local file.
Make sure the header validation is done before calling this.
Args:
bucket (str): S3 bucket
key (str): S3 key
file_path (str): Local file path to store the contents
start_range (int): Start range of ScanRange parameter of S3 Select
end_range (int): End range of ScanRange parameter of S3 Select
delimiter (str): S3 file delimiter
header_row (str): Header row of the local file. This will be inserted as first line in local file.
"""
aws_profile = current_app.config.get('AWS_PROFILE_NAME')
s3_client = boto3.session.Session(profile_name=aws_profile).client('s3')
expression = 'SELECT * FROM S3Object'
try:
response = s3_client.select_object_content(
Bucket=bucket,
Key=key,
ExpressionType='SQL',
Expression=expression,
InputSerialization={
'CSV': {
'FileHeaderInfo': 'USE',
'FieldDelimiter': delimiter,
'RecordDelimiter': '\n'
}
},
OutputSerialization={
'CSV': {
'FieldDelimiter': delimiter,
'RecordDelimiter': '\n',
},
},
ScanRange={
'Start': start_range,
'End': end_range
},
)
"""
select_object_content() response is an event stream that can be looped to concatenate the overall result set
"""
f = open(file_path, 'wb') # we receive data in bytes and hence opening file in bytes
f.write(header_row.encode())
f.write('\n'.encode())
for event in response['Payload']:
if records := event.get('Records'):
f.write(records['Payload'])
f.close()
except ClientError:
logger.exception(f'Client error reading S3 file {bucket} : {key}')
except Exception:
logger.exception(f'Error reading S3 file {bucket} : {key}')
That's it! 😎 Now, instead of streaming the S3 file bytes by bytes, we parallelize the processing by concurrently processing the chunks. It wasn't that tough, isn't it? 😅
🔍 Comparing the processing time
If we compare the processing time of the same file we processed in our last post with this approach, the processing runs approximately 68% faster (with the same hardware and config). 😆
| Streaming S3 File | Parallel Processing S3 File | |
|---|---|---|
| File size | 4.8MB | 4.8MB |
| Processing time | ~37 seconds | ~12 seconds |
✔️ Benefits of this approach
- A very large file containing millions of records can be processed within minutes. I have been using this approach in the production environment for a while, and it's very blissful
- Computing and processing is distributed among distributed workers
- Processing speed can be tweaked by the availability of worker pools
- No more memory issues
📌 You can check out my GitHub repository for a complete working example of this approach 👇
 idris-rampurawala
/
s3-select-demo
idris-rampurawala
/
s3-select-demo
This project showcases the rich AWS S3 Select feature to stream a large data file in a paginated style.
AWS S3 Select Demo
This project showcases the rich AWS S3 Select feature to stream a large data file in a paginated style.
Currently, S3 Select does not support OFFSET and hence we cannot paginate the results of the query. Hence, we use scanrange feature to stream the contents of the S3 file.
Background
Importing (reading) a large file leads Out of Memory error. It can also lead to a system crash event. There are libraries viz. Pandas, Dask, etc. which are very good at processing large files but again the file is to be present locally i.e. we will have to import it from S3 to our local machine. But what if we do not want to fetch and store the whole S3 file locally at once? 🤔
Well, we can make use of AWS S3 Select to stream a large file via it's ScanRange parameter. This approach…
📑 Resources
- My GitHub repository demonstrating the above approach
- AWS S3 Select boto3 reference
- AWS S3 Select userguide





Top comments (0)