How to convert from CSV to a Columnar format without using a cluster?
Converting from a flat format such as CSV or JSON to a columnar storage (such as ORC or Parquet is a task that any data engineer will have to do probably multiple times each week).
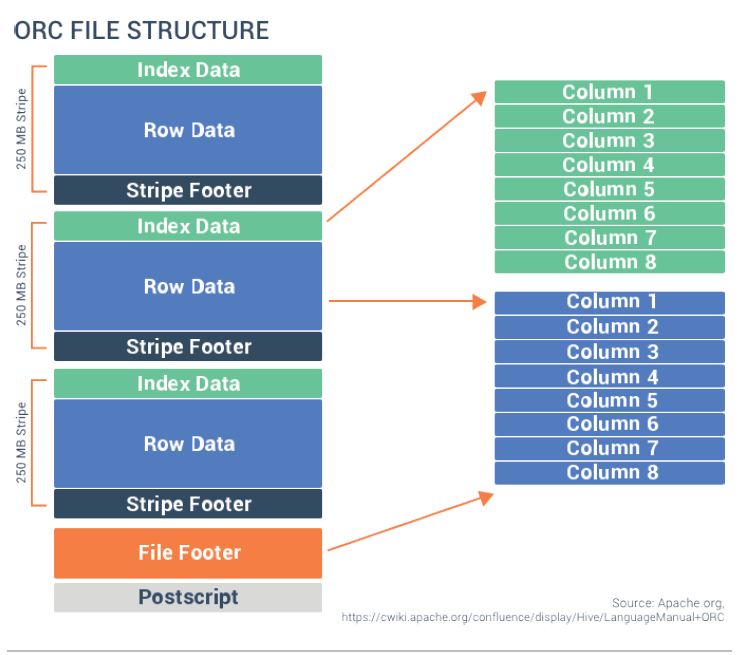
CSV's are row based (think Excel - you have Column names at the top and then the values in each row going down).
Columnar formats are different (See ORC Example below):
And the process of converting between a flat, row based format to a columnar format is computationally expensive that usually requires the creation of a cluster to manage this effectively in a reasonable amount of time.
Ways of converting CSV to ORC
I am a great believer in simplicity when it comes to writing files like this. In our previous experiments we realised that although using things like Hive can greatly simplify the process and allow it to be more automated. Systems like Spark can actually manage the process faster and cheaper if you keep things simple.
Hive / Presto
In a nutshell, you create a new table with STORED AS ORC on the definition. You then:
INSERT into orc_table SELECT * from csv_table
This then rewrites your CSV data into a new ORC table on the fly.
This requires a multi-node cluster to ensure things run quickly.
Dataframes
Spark runs on dataframes. You essentially load files into a dataframe and then output that dataframe as a different type of file.
csv = df.read_csv('/path/to/csv/files')
csv.to_orc('/path/for/final/orc/files')
I mean it doesn't really get any simpler than that for a 1-1 conversion. However this still requires a cluster to work and its speed is very much based on the number of nodes you point at it / how you manage node memory.
Enter the GPU
GPU processing used to be the domain of Game designers and some scientist types exclusively. However with the recent introduction of frameworks such as rapids.ai (https://rapids.ai) things have got much easier for the average Joe.
I recently set up my QNAP NAS as a data-science lab (you can read about the setup of it and Rapids.Ai here: https://www.linkedin.com/pulse/setting-up-rapidsai-qnap-nas-dan-voyce).
This included (naturally) an obscene amount of storage, some fast NVME hard-drives and most importantly a GPU (I went with a cheap 1060 GTX with 6GB memory as max speed isn't required for just a PoC).
Using Rapids.ai & cuDF to convert formats using the GPU.
As I have said - my setup is not particularly complex or the most up-to-date.
Mark Litwintschik is a fantastic Big Data consultant who transparently publishes comparisons of various methods of CSV conversion on his blog: https://tech.marksblogg.com/faster-csv-to-orc-conversions.html
In his comparisons he launches a 21 node m3.xlarge cluster on AWS to complete his conversions. According to the AWS cost calculator this costs $430 USD per month if used for a maximum of 2 hours per day (Marks longest conversion)
In comparison a single EC2 instance with a 8GB GPU runs at $54 USD per month for 2 hours usage per day (This is an Linux on i3en.xlarge which is probably overkill for the computing but includes 2500GB of NVME Storage)
There are other factors here that I haven't tested yet such as S3 latency and speed over Hard Drives but in my experience they are roughly on par.
By adding local NVME storage to the instance it means you can process this locally, however for truly huge datasets it is sometimes preferable to just process from S3 directly and output back to S3 - YMMV.
The Dataset
We were asked to produce a sample dataset for Las Vegas for a client. Generally unless clients are super tech savvy they request files in CSV format, unfortunately for us this is not the most efficient way of querying things so I figured a real-world dataset would be a good test of this.
The dataset comes in at just over 2 Billion Rows and has 41 fields. The total size of this data is 397GB uncompressed or around 127GB gzip compressed.
So this is a little larger than the dataset that Mark has done his benchmarks on but should be a good comparison.
A note on partitions:
I am using an older consumer graphics card that only has 6GB of memory.
Whatever Dask creates, the partitions still need to fit into the available GPU memory, obviously having 3000 files in this case is not ideal and whilst I could probably tune it to be less if having this number of files is a deal breaker for you then I would suggest maybe looking at a GPU with higher memory or attaching several GPU's to get a larger memory block. It should still work out much cheaper than a cluster!
First test on "Spinning Rust"
My NAS has 8 x 4TB Seagate Ironwolf SATA drives setup in RAID-10, this usually gets me around 140MB/s read speed.
On the Jupyter notebook that is provided with rapids.ai I ran the following:
The output is included
%%time
import dask_cudf as dc
ddf = dc.read_csv('/data/Data Files/Vegas/datafiles/csv/*.csv.gz', compression='gzip')
CPU times: user 1.82 s, sys: 870 ms, total: 2.69 s
Wall time 6.99 s
%%time
ddf = ddf.repartition(npartitions=3000)
CPU times: user 60.2 ms, sys: 159 µs, total: 60.4 ms
Wall time: 57.6 ms
%%time
ddf.to_orc('/data/Data Files/Vegas/datafiles/orc/')
CPU times: user 1h 4min 4s, sys: 30min 19s, total 1h 34min 23s
Wall time: 41min 57s
42 Minutes - wow!
These tests are already pretty mind blowing. The fact that my consumer NAS with its out of date consumer graphics card can beat the majority of the benchmarks for converting CSV data that Mark provided (that are run on huge clusters) is amazing. I have no doubt that with NVME drives this will be significantly faster.
First test on NVMe drive
I added a 1TB NVMe drive (I bought 2 but it seems like my QNAP doesnt want to recognise anything in NVMe Slot 1 (RMA incoming)), to my NAS and provisioned it as a volume. I copied the dataset to the newly provisioned drives (mounted at /ssd) and re-ran the test:
%%time
import dask_cudf as dc
ddf = dc.read_csv('/ssd/csv/*.csv.gz', compression='gzip')
CPU times: user 1.87 s, sys: 726 ms, total: 2.59 s
Wall time: 2.13 s
%%time
ddf = ddf.repartition(npartitions=3000)
CPU times: user 76 ms, sys: 22 ms, total: 98 ms
Wall time: 97.2 ms
%%time
ddf.to_orc('/ssd/orc/')
CPU times: user 1h 16min 7s, sys: 18min 50s, total: 1h 34min 57s
Wall time: 32min 15s
32 Minutes
This is obviously faster but a bit disappointing, I was expecting around the 20 minute benchmark, however this is likely because I am reading and writing to / from the same drive. Unfortunately with my NAS not recognising the other NVMe drive I can't really do a proper test until it is fixed.
CONCLUSIONS
Using a GPU is basically like using a mini-cluster, but rather instead of spreading the workload out onto multiple machines and therefore multiple CPU cores, it does it all within its own CUDA cores.
The fact that my ASUS GTX 1060 6GB card can do a basic conversion from Row based to Column based formats in less time than a 21 node, large instance cluster should be eye opening at the least.
I will also be testing out our spatial joins on GPU in another article to follow!

|
Daniel Voyce - CTO Daniel leads the technology strategy, architecture and development at LOCALLY.
|


Top comments (0)