I decided to go on an adventure. I will write a simple biological cell simulator in Rust. This is my first time to write a simulator and also my first time using Rust.
In the early stage it will be useful only for the better understanding of already known biological concepts. However, later hopefully it will help to experiment with evolutionary and pharmacological questions. My long term goal is to simulate multiple Escherichia coli bacteria in a complex environment. These bacteria are very well studied, thanks to their significance in human and animal health. They can be both beneficial as probiotics or harmful as pathogens.
I have choosen Rust, because it is a fast, low level language. It has comparable performance to C and C++, but unlike them it has a built-in and streamlined solution for package management and documentation generation. I was able to use them in 5 minutes as a first timer.
However the most telling indicator, that I was able to write the most simple proof of concept in around an hour. Somehow the language abstractions are similar to how I like to model problems in my head, which is deeply subjective, but still it strengthened my decision.
I plan to work in baby steps, so first I programmed a very simple diffusion model.
If you did not hear about diffusion, its basic explanation is easy. Given you have a water tank and you infuse it with oxygen or with some other soluble substance, then the particles of that substance can traverse freely in any direction.
So given time, they will be more or less dispersed in the water tank uniformly. However, if you divide the water tank with a membrane, then you will have the same result, but after a longer time period. The time delay will be dependent on the membrane permeability for that given substance. The lower the permeability, the longer time you must wait.
I wrote pseudo-code-like python script to provide the simplest model for this problem.
def membrane_permeate(a_solvent_o2_count, b_solvent_o2_count, o2_permeability):
o2_forward = int(round(a_solvent_o2_count * o2_permeability))
o2_reverse = int(round(b_solvent_o2_count * o2_permeability))
a_solvent_o2_count = a_solvent_o2_count - o2_forward
b_solvent_o2_count = b_solvent_o2_count - o2_reverse
a_solvent_o2_count = a_solvent_o2_count + o2_reverse
b_solvent_o2_count = b_solvent_o2_count + o2_forward
return a_solvent_o2_count, b_solvent_o2_count
a_solvent_o2_count = 10000
b_solvent_o2_count = 10
o2_permeability = 0.3
for cycle in range(10):
a_solvent_o2_count, b_solvent_o2_count = membrane_permeate(
a_solvent_o2_count,
b_solvent_o2_count,
o2_permeability,
)
print(f"{cycle},{a_solvent_o2_count},{b_solvent_o2_count}")
The Rust implementation logically is almost the same, but for 3 equal volume solvent spaces across 2 membranes between them.
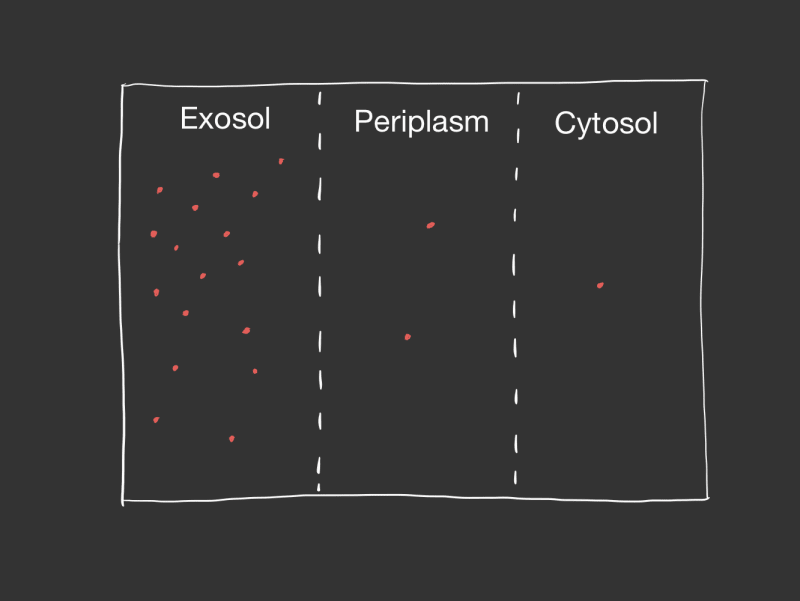
Well it is almost a gram negative bacteria, only a few parts are missing. XD
Also you should note that I came up with the term "exosol". So on your quiz you should write what your teacher said, which will be most likely "extracellular environment" which is long and boring but will be the right answer.
Back to the topic: I fell in love with rust, because with the trait language feature I was able to program diffusion in a very close way to how I would reason about it:
"Diffusion is based on the trait of membranes that they can permeate molecules between solvents, which they separate"
and one of the most important attributes of every code is how easy it is to reason about it.
So let's see the code:
First I defined the Solvent struct, which has only one field, the oxygen molecule count. Rust structs seems very similar to C structs, but differ in some important ways. The most important difference is regarding this code is the ability in Rust to implement functions bound to a struct.
struct Solvent {
o2_count: i128,
}
Then I defined the Membrane also as a struct, has a sole field, its permeability for molecular oxygen, expressed as a simple factor.
struct Membrane {
o2_permeability: f32,
}
Then I defined the Permeate trait, which has a method with 2 solvents as it parameters.
trait Permeate {
fn permeate(&self, solvent1: &mut Solvent, solvent2: &mut Solvent);
}
Then I implemented the Permeate trait for the Membrane struct. I just calculated the amount of oxygen flux in both directions by multiplying the oxygen count in the given simulation cycle with the membrane permeability factor.
impl Permeate for Membrane {
fn permeate(&self, solvent1: &mut Solvent, solvent2: &mut Solvent) {
let o2_forward = (solvent1.o2_count as f32) * self.o2_permeability;
let o2_reverse = (solvent2.o2_count as f32) * self.o2_permeability;
solvent1.o2_count = solvent1.o2_count - (o2_forward.round() as i128);
solvent2.o2_count = solvent2.o2_count - (o2_reverse.round() as i128);
solvent1.o2_count = solvent1.o2_count + (o2_reverse.round() as i128);
solvent2.o2_count = solvent2.o2_count + (o2_forward.round() as i128);
}
}
Lastly in the main function I declare the players: extrasol, periplasm, cytosol, outer_membrane and inner_membrane. So in a simulation loop I can calculate the diffusion across the two membranes.
The most important two lines are:
outer_membrane.permeate(&mut extrasol, &mut periplasm);
inner_membrane.permeate(&mut periplasm, &mut cytosol);
That is all it takes to calculate the diffusion in every simulation cycle. It is so tidy.
However here is the full main function:
fn main() {
let mut extrasol = Solvent { o2_count: 10000 };
let mut periplasm = Solvent { o2_count: 10 };
let mut cytosol = Solvent { o2_count: 5 };
let outer_membrane = Membrane {
o2_permeability: 0.3,
};
let inner_membrane = Membrane {
o2_permeability: 0.3,
};
println!("cycle,extrasol_o2,periplasm_o2,cytosol_o2");
for cycle in 1..=10 {
println!(
"{},{},{},{}",
cycle, extrasol.o2_count, periplasm.o2_count, cytosol.o2_count
);
outer_membrane.permeate(&mut extrasol, &mut periplasm);
inner_membrane.permeate(&mut periplasm, &mut cytosol);
}
}
So it was a nice experience. In the next version I will step up my game by developing a more sophisticated diffusion model, because now I am very far away from a strong model. One of the most obvious limitations is that in reality those solvents have vastly different volumes. Also the membrane surface areas should be considered. So I will further develop the model using Fick's law. Which I will explain in the next post.
But nevertheless I have a very simple model as a starting point on my way to a nicer bacteria model and I also learnt a bit of Rust, which was joyful.
This project is free and open source under GPL 3.0 license,and can be found on GitHub.









Top comments (0)