After trying to read J.R.R. Tolkien's The Silmarillion again for the millionth time, I remembered a funny tweet that has been around for a while:
Even though I was a casual fan of The Lord of the Rings and having already taken two pharmacology courses in college, I had no idea who or what a Narmacil was. Should we fear him/her by its sword skills or by its dangerous side effects?
This little trivia prompted me to ask if an artificial neural network (ANN) could succeed where I and many more have failed. Here, I show you how to build a special type of ANN called Long Short-Term Memory (LSTM) to classify Tolkien characters and prescription drug names using Keras.
Dataset
The first step was to build from scratch a combined dataset with names of Tolkien characters and prescription drugs (a bunch of them, not just antidepressants).
Tolkien characters
Lucky for us, the Behind the Name website has a database of the first names of Tolkien characters that we can directly read from the page's HTML using pandas.
import pandas as pd
raw_tolkien_chars = pd.read_html('https://www.behindthename.com/namesakes/list/tolkien/name')
raw_tolkien_chars[2].head()
| Name | Gender | Details | Total | |
|---|---|---|---|---|
| 0 | Adalbert | m | 1 character | 1 |
| 1 | Adaldrida | f | 1 character | 1 |
| 2 | Adalgar | m | 1 character | 1 |
| 3 | Adalgrim | m | 1 character | 1 |
| 4 | Adamanta | f | 1 character | 1 |
tolkien_names = raw_tolkien_chars[2]['Name']
tolkien_names.iloc[350:355]
350 Gethron
351 Ghân-buri-Ghân
352 Gildis
353 Gildor
354 Gil-galad
Name: Name, dtype: object
We can see that some names are hyphenated and have accented letters. To simplify the analysis I transformed unicode characters to ASCII, removed punctuation marks, transformed them to lowercase, and removed any possible duplicates.
import unidecode
processed_tolkien_names = tolkien_names.apply(unidecode.unidecode).str.lower().str.replace('-', ' ')
processed_tolkien_names = [name[0] for name in processed_tolkien_names.str.split()]
processed_tolkien_names = pd.DataFrame(processed_tolkien_names, columns=['name']).sort_values('name').drop_duplicates()
processed_tolkien_names['tolkien'] = 1
processed_tolkien_names['name'].iloc[350:355]
473 gethron
439 ghan
109 gil
341 gildis
324 gildor
Name: name, dtype: object
processed_tolkien_names.shape
(746,2)
Done! Now we have 746 different character names.
Prescription drugs
To get a comprehensive list of drug names, I downloaded the medication guide of the U.S. Food & Drug Administration (FDA).
raw_medication_guide = pd.read_csv('data/raw/medication_guides.csv')
raw_medication_guide.head()
| Drug Name | Active Ingredient | Form;Route | Appl. No. | Company | Date | Link | |
|---|---|---|---|---|---|---|---|
| 0 | Abilify | Aripiprazole | TABLET, ORALLY DISINTEGRATING;ORAL | 21729 | OTSUKA | 02/05/2020 | https://www.accessdata.fda.gov/drugsatfda_docs... |
| 1 | Abilify | Aripiprazole | TABLET;ORAL | 21436 | OTSUKA | 02/05/2020 | https://www.accessdata.fda.gov/drugsatfda_docs... |
| 2 | Abilify | Aripiprazole | SOLUTION;ORAL | 21713 | OTSUKA | 02/05/2020 | https://www.accessdata.fda.gov/drugsatfda_docs... |
| 3 | Abilify | Aripiprazole | SOLUTION;ORAL | 21713 | OTSUKA | 02/05/2020 | https://www.accessdata.fda.gov/drugsatfda_docs... |
| 4 | Abilify | Aripiprazole | INJECTABLE;INTRAMUSCULAR | 21866 | OTSUKA | 02/05/2020 | https://www.accessdata.fda.gov/drugsatfda_docs... |
drug_names = raw_medication_guide['Drug Name']
drug_names.iloc[160:165]
160 Chantix
161 Children's Cetirizine Hydrochloride Allergy
162 Chlordiazepoxide and Amitriptyline Hydrochloride
163 Cimzia
164 Cimzia
Name: Drug Name, dtype: object
A similar preprocessing step was repeated for this dataset too:
processed_drug_names = drug_names.str.lower().str.replace('.', '').str.replace( '-', ' ').str.replace('/', ' ').str.replace("'", ' ').str.replace(",", ' ')
processed_drug_names = [name[0] for name in processed_drug_names.str.split()]
processed_drug_names = pd.DataFrame(processed_drug_names, columns=['name']).sort_values('name').drop_duplicates()
processed_drug_names['tolkien'] = 0
processed_drug_names['name'].iloc[84:89]
373 chantix
448 children
395 chlordiazepoxide
185 cimzia
292 cipro
Name: name, dtype: object
processed_drug_names.shape
(611,2)
Done, 611 different drug names!
We can finally combine the two datasets and move on.
dataset = pd.concat([processed_tolkien_names, processed_drug_names], ignore_index=True)
Data transformation
So now we have a bunch of names, but machine learning models don't work with raw characters. We need to convert them into a numerical format that can be processed by our soon-to-be-built model.
Using the Tokenizer class from Keras, we set char_level=True to process each word at character-level. The fit_on_texts() method will update the tokenizer internal vocabulary based on our dataset names and then texts_to_sequences() will transform each name into a sequence of integers.
from keras.preprocessing.text import Tokenizer
tokenizer = Tokenizer(char_level=True)
tokenizer.fit_on_texts(dataset['name'])
char_index = tokenizer.texts_to_sequences(dataset['name'])
Look how our beloved Bilbo is now:
print(dataset['name'][134])
print(char_index[134])
bilbo
[16, 3, 6, 16, 5]
Yet, this representation is not ideal. Having integers to represent letters could lead the ANN to assume that the characters have an ordinal scale. To solve this problem we have to:
Set all names to have the length of the longest name (17 characters here). We use
pad_sequencesto add 0's to the end of names shorter than 17 letters.Convert each integer representation to its one-hot encoded vector representation. The vector consists of 0s in all cells except for a single 1 in a cell to identify the letter.
from keras.preprocessing.sequence import pad_sequences
from keras.utils import to_categorical
import numpy as np
char_index = pad_sequences(char_index, maxlen=dataset['name'].apply(len).max(), padding="post")
x = to_categorical(char_index) # onehot encoding
y = np.array(dataset['tolkien'])
x.shape
(1357, 17, 27)
We have 1357 names. Each name has 17 letters and each letter is a one-hot encoded vector of size 27 (26 letters of the Latin alphabet + padding character).
Data split
I split the data into train, validation, and test sets with a 60/20/20 ratio using a custom function since sklearn train_test_split only outputs two sets.
from sklearn.model_selection import train_test_split
def data_split(data, labels, train_ratio=0.5, rand_seed=42):
x_train, x_temp, y_train, y_temp = train_test_split(data,
labels,
train_size=train_ratio,
random_state=rand_seed)
x_val, x_test, y_val, y_test = train_test_split(x_temp,
y_temp,
train_size=0.5,
random_state=rand_seed)
return x_train, x_val, x_test, y_train, y_val, y_test
x_train, x_val, x_test, y_train, y_val, y_test = data_split(x, y, train_ratio=0.6)
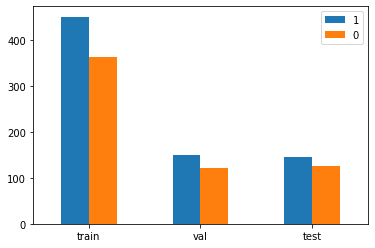
Let's take a look at the splits:
from collections import Counter
import matplotlib.pyplot as plt
dataset_count = pd.DataFrame([Counter(y_train), Counter(y_val), Counter(y_test)],
index=["train", "val", "test"])
dataset_count.plot(kind='bar')
plt.xticks(rotation=0)
plt.show()
print(f"Total number of samples: \n{dataset_count.sum(axis=0).sum()}")
print(f"Class/Samples: \n{dataset_count.sum(axis=0)}")
print(f"Split/Class/Samples: \n{dataset_count}")
Total number of samples:
1357
Class/Samples:
1 746
0 611
dtype: int64
Split/Class/Samples:
1 0
train 451 363
val 149 122
test 146 126
There are more Tolkien characters than drug names, but it seems like a decent balance.
LSTM model
Long Short-Term Memory is a type of Recurrent Neural Network proposed by Hochreiter S. & Schmidhuber J. (1997) to store information over extended time intervals. Names are just sequences of characters in which the order is important, so LSTM networks are a great choice for our name prediction task. You can read more about LSTMs in this awesome illustrated guide written by Michael Phi.
Training
Keras was used to build this simple LSTM model after some tests and hyperparameter tuning. It is just a hidden layer with 8 LSTM blocks, one dropout layer to prevent overfitting, and one output neuron with a sigmoid activation function to make a binary classification.
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense, Activation, Dropout, LSTM
from tensorflow.random import set_seed
set_seed(23)
model = Sequential()
model.add(LSTM(8, return_sequences=False,
input_shape=(x.shape[1], x.shape[2])))
model.add(Dropout(0.3))
model.add(Dense(units=1))
model.add(Activation('sigmoid'))
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
lstm (LSTM) (None, 8) 1152
_________________________________________________________________
dropout (Dropout) (None, 8) 0
_________________________________________________________________
dense (Dense) (None, 1) 9
_________________________________________________________________
activation (Activation) (None, 1) 0
=================================================================
Total params: 1,161
Trainable params: 1,161
Non-trainable params: 0
_________________________________________________________________
Adam is a good default optimizer and produces great results in deep learning applications. Binary cross-entropy is the default loss function to binary classification problems and it is compatible with our single neuron output architecture.
from tensorflow.keras.optimizers import Adam
model.compile(loss="binary_crossentropy",
optimizer=Adam(learning_rate=1e-3), metrics=['accuracy'])
Two callbacks were implemented. EarlyStopping to stop the training process after 20 epochs without reducing the validation loss and ModelCheckpoint to always save the model when the validation loss drops.
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
es = EarlyStopping(monitor='val_loss', verbose=1, patience=20)
mc = ModelCheckpoint("best_model.h5", monitor='val_loss',
verbose=1, save_best_only=True)
history = model.fit(x_train, y_train, batch_size=32, epochs=100,
validation_data=(x_val, y_val), callbacks=[es, mc])
Epoch 00071: val_loss did not improve from 0.34949
Epoch 72/100
26/26 [==============================] - 1s 24ms/step - loss: 0.3085 - accuracy: 0.8836 - val_loss: 0.3861 - val_accuracy: 0.8487
Epoch 00072: val_loss did not improve from 0.34949
Epoch 00072: early stopping
val_loss_per_epoch = history.history['val_loss']
best_epoch = val_loss_per_epoch.index(min(val_loss_per_epoch)) + 1
print(f"Best epoch: {best_epoch}")
Best epoch: 52
Let's plot the accuracy and loss values per epoch to see the progression of these metrics.
def plot_metrics(history):
plt.figure(figsize=(12,6))
plt.subplot(1,2,1)
plt.plot(history.history['accuracy'], label='Training')
plt.plot(history.history['val_accuracy'], label='Validation')
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
plt.grid('on')
plt.subplot(1,2,2)
plt.plot(history.history['loss'], label='Training')
plt.plot(history.history['val_loss'], label='Validation')
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend(loc='upper right')
plt.grid('on')
plot_metrics(history)
We can see that the accuracy quickly reaches a good plateau around 80%. Visually the model appears to start overfitting after epoch 50. It shouldn't be a problem to use the version saved by ModelCheckpoint at epoch 52.
Performance evaluation
Finally, let's see how our model does with the test dataset.
from tensorflow.keras.models import load_model
model = load_model("best_model.h5")
metrics = model.evaluate(x=x_test, y=y_test)
9/9 [==============================] - 1s 7ms/step - loss: 0.4595 - accuracy: 0.8125
print("Accuracy: {0:.2f} %".format(metrics[1]*100))
Accuracy: 81.25 %
81.25 %
Not bad!
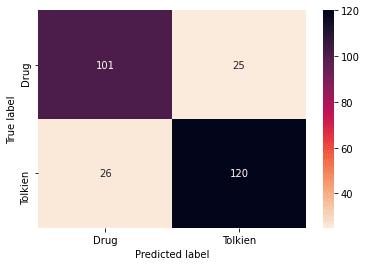
We can explore the results a little more with the confusion matrix and classification report:
from sklearn.metrics import confusion_matrix
from seaborn import heatmap
def plot_confusion_matrix(y_true, y_pred, labels):
cm = confusion_matrix(y_true, y_pred)
heatmap(cm, annot=True, fmt="d", cmap="rocket_r", xticklabels=labels, yticklabels=labels)
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt.show()
predictions = model.predict(x_test)
threshold = 0.5
y_pred = predictions > threshold
plot_confusion_matrix(y_test, y_pred, labels=['Drug','Tolkien'])
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred, target_names=['Drug', 'Tolkien']))
precision recall f1-score support
Drug 0.80 0.80 0.80 126
Tolkien 0.83 0.82 0.82 146
accuracy 0.81 272
macro avg 0.81 0.81 0.81 272
weighted avg 0.81 0.81 0.81 272
Besides the good accuracy, the model has almost the same number of false positives and false negatives. We can see this reflecting in a balanced precision and recall.
So, just out of curiosity, which Tolkien characters could pass as prescription drugs?
def onehot_to_text(onehot_word):
"""Reverse one-hot encoded words to strings"""
char_index = [[np.argmax(char) for char in onehot_word]]
word = tokenizer.sequences_to_texts(char_index)
return ''.join(word[0].split())
test_result = pd.DataFrame()
test_result['true'] = y_test
test_result['prediction'] = y_pred.astype(int)
test_result['name'] = [onehot_to_text(name) for name in x_test]
test_result.head()
| true | prediction | name | |
|---|---|---|---|
| 0 | 0 | 0 | supprelin |
| 1 | 1 | 1 | bingo |
| 2 | 0 | 0 | ponstel |
| 3 | 0 | 1 | elidel |
| 4 | 0 | 0 | aubagio |
test_result['name'].loc[(test_result['true']==1) & (test_result['prediction']==0)]
13 ivy
17 camellia
44 celebrindor
47 meriadoc
63 vanimelde
64 finduilas
75 eglantine
84 ruby
87 poppy
89 otto
100 tanta
102 myrtle
108 prisca
132 cottar
151 stybba
171 este
175 daisy
189 tulkas
195 arciryas
205 odovacar
206 tarcil
207 hyarmendacil
229 jago
230 tata
240 ponto
271 landroval
Name: name, dtype: object
Conclusion
So, here we covered how to work with character embeddings and build a simple LSTM model capable of telling apart Tolkien character names from prescription drug names. Full code, including requirements, dataset, a Jupyter Notebook code version, and a script version, can be found at my GitHub repo.
You can also play around with this popular interactive quiz found on the web: Antidepressant or Tolkien?. I only got 70.8% right! Can you guess better than the LSTM network?
References
Hu, Y., Hu, C., Tran, T., Kasturi, T., Joseph, E., & Gillingham, M. (2021). What's in a Name?--Gender Classification of Names with Character Based Machine Learning Models. arXiv preprint arXiv:2102.03692.
Bhagvati, C. (2018). Word representations for gender classification using deep learning. Procedia computer science, 132, 614-622.
Liang, X. (2018). How to Preprocess Character Level Text with Keras.
















Top comments (1)
This is awesome -- and I LOVE all of The Lord of the Rings references!